Followers of this blog and our team’s scientific endeavors may know we have a curated database of brown dwarfs we work with. An initial version of this database has been published in Filippazzo et al. 2015 and contains information for 198 objects. The database is also maintained on Github, where we welcome contributions from other researchers. We’ve developed a set of tools for astronomers to work with SQL databases, namely the Python package astrodbkit. This package can be applied to other SQL databases allowing astronomers from all fields of research to manage their data.
Here we introduce a new tool: AstrodbWeb, a web-based interface to explore the BDNYC database.
Continue reading
Tag Archives: database
Creating a Database with astrodbkit
NOTE: The information on this post may be outdated. We recommend looking into the up-to-date documentation at trac (internal) and ReadTheDocs.
The astrodbkit package can be used to modify an existing SQL database (such as The BDNYC Database) but it can also be used to create and populate a SQL database from scratch.
To do this, import the BDdb module and create a new database with
from astrodbkit import astrodb
dbpath = '/path/to/new_database.db'
astrodb.create_databse(dbpath)
Then load your new database with
db = astrodb.Database(dbpath)
and start adding tables! The db.table() method accepts as its arguments the table name, list of field names, and list of data types like so:
db.table('my_new_table', ['field1','field2'], ['INTEGER','TEXT'], new_table=True)
Note new_table=True is necessary to create a new table. Otherwise, it looks for an existing table to modify (which you could do as well!).
To populate your new database with data, read the documentation here or a summary at Adding Data to the BDNYC Database.
As always, I recommend the SQLite Browser for a nice GUI to make changes outside of the command line.
Happy databasing!
Updating the astrodbkit documentation
As you make changes to the astrodbkit repository on Github, you may find that the documentation needs updating. Luckily, we use the invaluable Sphinx and the awesome ReadTheDocs to generate the documentation so this is fairly simple.
First, make sure you update the appropriate doc strings (those informative green bits just below the function definition) as this is what Sphinx will use to generate the documentation!
Start in the top level astrodbkit directory and generate the documentation from the module docstrings with
sphinx-build docs astrodbkit -a
Then cd into the docs directory and rebuild the html files with
cd docs
make html
Now move back to the top level directory and add, commit, and push your changes to Github with something like
cd ..
git add /docs
git commit -m "Updated the documentation for methods x, y, and z."
git push origin <your_branch>
All set! Refresh the page (after a few minutes so ReadTheDocs can build the pages) and make sure everything is to your liking. Well done.
And just in case, here’s a great tutorial for getting started with Sphinx and here’s the official documentation.
Deploying Package Releases to PyPI
Registering the package
To register the package and deploy the initial release to PyPI, do:
python setup.py register -r pypi
python setup.py sdist upload -r pypi
Deploying a new release
So you’ve made some cool new improvements to your Python package and you want to deploy a new release. It’s just a few easy steps.
- After your package modules have been updated, you have to update the version number in your setup.py file. The format is major.minor.micro depending on what you changed. For example, a small bug fix to v0.2.3 would increment to v0.2.4 while a new feature might increment to v0.3.0.
- Then make sure all the changes are committed and pushed to Github.
- Build with
python setup.py sdist - Twine with
twine upload dist/compressed_package_filename
That’s it!
Adding Data to a SQL Database using astrodbkit
NOTE: The information on this post may be outdated. We recommend looking into the up-to-date documentation at trac (internal) and ReadTheDocs.
To add data to any table, there are two easy steps. As our working example, we’ll add some new objects to the SOURCES table.
Step 1: Create an ascii file of the data
First, you must choose a delimiter, which is just the character that will break up the data into columns. I recommend a pipe ‘|’ character since they don’t normally appear in text. This is better than a comma since some data fields may have comma-separated values.
Put the data to be added in an ascii file with the following formatting:
- The first line must be the |-separated column names to insert/update, e.g.
ra|dec|publication_id. Note that the column names in the ascii file need not be in the same order as the table. Also, only the column names that match will be added and non-matching or missing column names will be ignored, e.g.spectral_type|ra|publication_id|decwill ignore thespectral_typevalues as this is not a column in the SOURCES table and input the other columns in the correct places. - If a record (i.e. a line in your ascii file) has no value for a particular column, type nothing. E.g. for the given column names
ra|dec|publication_id|comments, a record with nopublication_idshould read34.567|12.834||This object is my favorite!.
Step 2: Add the data to the specified table
To add the data to the table (in our example, the SOURCES table), import astrodbkit and initialize the .db file. Then run the add_data() method with the path to the ascii file as the first argument and the table to add the data to as the second argument. Be sure to specify your delimiter with delim='|'. Here’s what that looks like:
from astrodbkit import astrodb
db = astrodb.Database('/path/to/the/database/file.db')
db.add_data('/path/to/the/upload/file.csv', 'sources', delim='|')
That’s it!
BDNYC Database Setup
WARNING: The information on this is outdated. We recommend looking into the up-to-date documentation at trac (internal) and ReadTheDocs.
Read this post first if your development environment needs setting up. Then…
Setting Up the Database
To install the database, open a Terminal session and do:
pip install BDNYCdb
Then download the public database file BDNCY198.db here or get the unpublished BDNYC.db file from Dropbox. (Ask a group member for the invite!)
Accessing the Database
Now you can load the entire database into a Python variable simply by launching the Python interpreter and pointing the get_db() function to the database file by doing:
In [1]: from BDNYCdb import BDdb
In [2]: db = BDdb.get_db('/path/to/your/Dropbox/BDNYCdb/BDNYC.db')
Voila!
In the database, each source has a unique identifier (or ‘primary key’ in SQL speak). To see an inventory of all data for one of these sources, just do:
In [3]: db.inventory(id)
where id is the source_id.
Now that you have the database at your fingertips, you’ll want to get some info out of it. To do this, you can use SQL queries.
Here is a detailed post about how to write an SQL query.
Further documentation for sqlite3 can be found here. Most commands involve wrapping SQL language inside python functions. The main method we will use to fetch data from the database is list():
In [5]: data = db.list( "SQL_query_goes_here" ).fetchall()
Example Queries
Some SQL query examples to put in the command above (wrapped in quotes of course):
- SELECT shortname, ra, dec FROM sources WHERE (222<ra AND ra<232) AND (5<dec AND dec<15)
- SELECT band, magnitude, magnitude_unc FROM photometry WHERE source_id=58
- SELECT source_id, band, magnitude FROM photometry WHERE band=’z’ AND magnitude<15
- SELECT wavelength, flux, unc FROM spectra WHERE observation_id=75”
As you hopefully gathered:
- Returns the shortname, ra and dec of all objects in a 10 square degree patch of sky centered at RA = 227, DEC = 10
- Returns all the photometry and uncertainties available for object 58
- Returns all objects and z magnitudes with z less than 15
- Returns the wavelength, flux and uncertainty arrays for all spectra of object 75
The above examples are for querying individual tables only. We can query from multiple tables at the same time with the JOIN command like so:
- SELECT t.name, p.band, p.magnitude, p.magnitude_unc FROM telescopes as t JOIN photometry AS p ON p.telescope_id=t.id WHERE p.source_id=58
- SELECT p1.magnitude-p2.magnitude FROM photometry AS p1 JOIN photometry AS p2 ON p1.source_id=p2.source_id WHERE p1.band=’J’ AND p2.band=’H’
- SELECT src.designation, src.unum, spt.spectral_type FROM sources AS src JOIN spectral_types AS spt ON spt.source_id=src.id WHERE spt.spectral_type>=10 AND spt.spectral_type<20 AND spt.regime=’optical’
- SELECT s.unum, p.parallax, p.parallax_unc, p.publication_id FROM sources as s JOIN parallaxes AS p ON p.source_id=s.id
As you may have gathered:
- Returns the survey, band and magnitude for all photometry of source 58
- Returns the J-H color for every object
- Returns the designation, U-number and optical spectral type for all L dwarfs
- Returns the parallax measurements and publications for all sources
Alternative Output
As shown above, the result of a SQL query is typically a list of tuples where we can use the indices to print the values. For example, this source’s g-band magnitude:
In [9]: data = db.list("SELECT band,magnitude FROM photometry WHERE source_id=58").fetchall()
In [10]: data
Out[10]: [('u', 25.70623),('g', 25.54734),('r', 23.514),('i', 21.20863),('z', 18.0104)]
In [11]: data[1][1]
Out[11]: 25.54734
However we can also query the database a little bit differently so that the fields and records are returned as a dictionary. Instead of db.query.execute() we can do db.dict() like this:
In [12]: data = db.dict("SELECT * FROM photometry WHERE source_id=58").fetchall()
In [13]: data
Out[13]: [<sqlite3.Row at 0x107798450>, <sqlite3.Row at 0x107798410>, <sqlite3.Row at 0x107798430>, <sqlite3.Row at 0x1077983d0>, <sqlite3.Row at 0x1077982f0>]
In [14]: data[1]['magnitude']
Out[14]: 25.54734
Database Schema and Browsing
In order to write the SQL queries above you of course need to know what the names of the tables and fields in the database are. One way to do this is:
In [15]: db.list("SELECT sql FROM sqlite_master").fetchall()
This will print a list of each table, the possible fields, and the data type (e.g. TEXT, INTEGER, ARRAY) for that field.
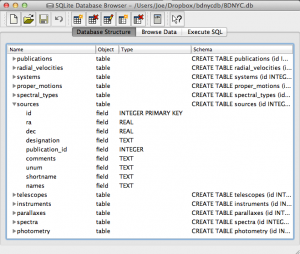 Even easier is to use the DB Browser for SQLite pictured at left which lets you expand and collapse each table, sort and order columns, and other fun stuff.
Even easier is to use the DB Browser for SQLite pictured at left which lets you expand and collapse each table, sort and order columns, and other fun stuff.
It even allows you to manually create/edit/destroy records with a very nice GUI.
IMPORTANT: If you are using the private database keep in mind that if you change a database record, you immediately change it for everyone since we share the same database file on Dropbox. Be careful!
Always check and double-check that you are entering the correct data for the correct source before you save any changes with the SQLite Database Browser.
SQL Queries
An SQL database is comprised of a bunch of tables (kind of like a spreadsheet) that have fields (column names) and records (rows of data). For example, our database might have a table called students that looks like this:
| id | first | last | grade | GPA |
|---|---|---|---|---|
| 1 | Al | Einstein | 6 | 2.7 |
| 2 | Annie | Cannon | 6 | 3.8 |
| 3 | Artie | Eddington | 8 | 3.2 |
| 4 | Carlie | Herschel | 8 | 3.2 |
So in our students table, the fields are [id, first, last, grade, GPA], and there are a total of four records, each with a required yet arbitrary id in the first column.
To pull these records out, we tell SQL to SELECT values for the following fields FROM a certain table. In SQL this looks like:
In [1]: db.execute("SELECT id, first, last, grade, GPA FROM students").fetchall()
Out[1]: [(1,'Al','Einstein',6,2.7),(2,'Annie','Cannon',6,3.8),(3,'Artie','Eddington',8,3.2),(4,'Carlie','Herschel',8,3.2)]
Or equivalently, we can just use a wildcard “*” if we want to return all fields with the SQL query "SELECT * FROM students".
We can modify our SQL query to change the order of fields or only return certain ones as well. For example:
In [2]: db.execute("SELECT last, first, GPA FROM students").fetchall()
Out[1]: [('Einstein','Al',2.7),('Cannon','Annie',3.8),('Eddington','Artie',3.2),('Herschel','Carlie',3.2)]
Now that we know how to get records from tables, we can restrict which records it returns with the WHERE statement:
In [3]: db.execute("SELECT last, first, GPA FROM students WHERE GPA>3.1").fetchall()
Out[3]: [('Cannon','Annie',3.8),('Eddington','Artie',3.2),('Herschel','Carlie',3.2)]
Notice the first student had a GPA less than 3.1 so he was omitted from the result.
Now let’s say we have a second table called quizzes which is a table of every quiz grade for all students that looks like this:
| id | student_id | quiz_number | score |
|---|---|---|---|
| 1 | 1 | 3 | 89 |
| 2 | 2 | 3 | 96 |
| 3 | 3 | 3 | 94 |
| 4 | 4 | 3 | 92 |
| 5 | 1 | 4 | 78 |
| 6 | 3 | 4 | 88 |
| 7 | 4 | 4 | 91 |
Now if we want to see only Al’s grades, we have to JOIN the tables ON some condition. In this case, we want to tell SQL that the student_id (not the id) in the quizzes table should match the id in the students table (since only those grades are Al’s). This looks like:
In [4]: db.execute("SELECT quizzes.quiz_number, quizzes.score FROM quizzes JOIN students ON students.id=quizzes.student_id WHERE students.last='Einstein'").fetchall()
Out[4]: [(3,89),(4,78)]
So students.id=quizzes.student_id associates each quiz with a student from the students table and students.last='Einstein' specifies that we only want the grades from the student with last name Einstein.
Similarly, we can see who scored 90 or greater on which quiz with:
In [5]: db.execute("SELECT students.last, quizzes.quiz_number, quizzes.score FROM quizzes JOIN students ON students.id=quizzes.student_id WHERE quizzes.score>=90").fetchall()
Out[5]: [('Cannon',3,96),('Eddington',3,94),('Herschel',3,92),('Herschel',4,91)]
That’s it! We can JOIN as many tables as we want with as many restrictions we need to pull out data in the desired form.
This is powerful, but the queries can become lengthy. A slight shortcut is to use the AS statement to assign a table to a variable (e.g. students => s, quizzes => q) like such:
In [6]: db.execute("SELECT s.last, q.quiz_number, q.score FROM quizzes AS q JOIN students AS s ON s.id=q.student_id WHERE q.score>=90").fetchall()
Out[6]: [('Cannon',3,96),('Eddington',3,94),('Herschel',3,92),('Herschel',4,91)]
